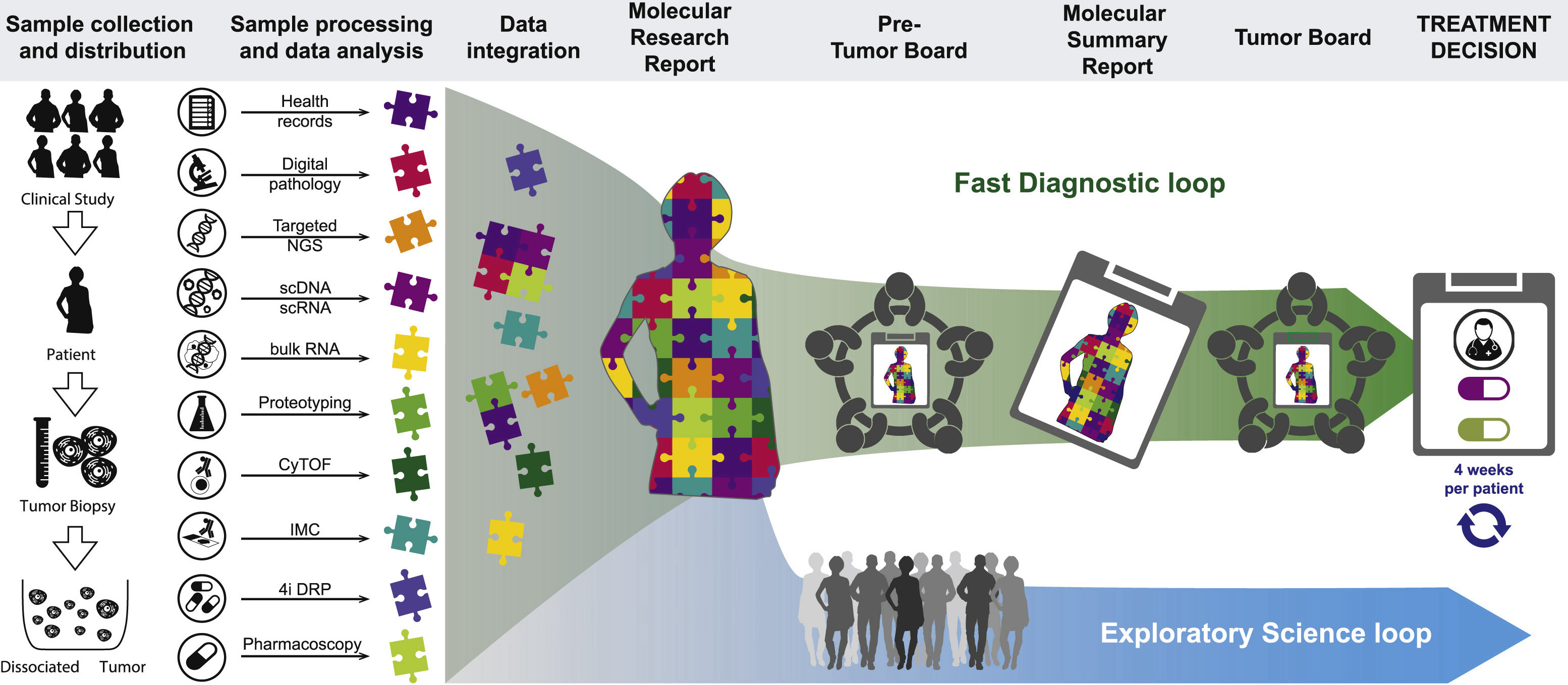
Tumor Profiler (TuPro) is an approved, observational clinical study (BASEC: 2018-02050, 2018-02052, 2019-01326) in which tumor samples are prospectively profiled and assessed whether combined multi-omics and functional readouts can provide evidence to support clinical decision-making beyond available and emerging diagnostic technologies such as digital pathology and targeted NGS. The technologies included in TuPro have been selected based on their ability to provide part of a multi-level depiction of the tumor or its microenvironment as well as their potential to deliver robust and clinically relevant insights in short turnaround times. The technologies are applied to 240 tumor samples collected over 3 years across three cancer indications: metastatic melanoma, metastatic epithelial ovarian cancer, and acute myeloid leukemia (AML). The selection of these indications is based on the potential clinical benefit and availability of sufficient tumor material for simultaneous analysis across all technologies. In addition to multiple bulk approaches, an average of two million single cells per patient are profiled across six technologies with single-cell readouts. The resulting data are analyzed immediately in the context of a "Fast Diagnostic loop," where their relevance to generate treatment recommendations on a per-patient basis is investigated. An in-depth analysis of the data acquired at the cohort level, including the clinical outcome of each patient collected over a 6-month follow-up period, is performed in the context of an "Exploratory Science loop," in which the multiscale approach will be used to improve the understanding of the disease and discover novel biomarkers.
TuPro includes two emerging clinical diagnostic approaches, i.e., tests that are not yet standard in cancer diagnostics, and seven exploratory profiling technologies in the TuPro study. Single-cell genomics approaches (scRNA [Papalexi and Satija, 2018] and scDNA [Kuipers et al., 2020]) generate a high-resolution map of the tumor microenvironment, characterize tumor cell heterogeneity, establish each tumor's evolutionary history, and take advantage of insights into cancer genomics and transcriptomics acquired over the past decades. In addition, bulk (DIA-MS) proteotyping (Gillet et al., 2012; Xuan et al., 2020) and single-cell CyTOF (Wagner et al., 2019) protein-based analyses are performed not only to expand on and translate transcriptomic observations but also to assess post-translational modifications affecting proteins involved in signaling pathways. The characterization of the tumor microenvironment is enriched with spatially resolved approaches: digital pathology and imaging mass cytometry (IMC) (Giesen et al., 2014) enable the characterization of cell-cell interactions within the tumor microenvironment by providing quantitative, single-cell, and spatially resolved data. This is of particular value for predicting the success of therapies that depend on direct cell-cell interactions, such as immune checkpoint inhibitors. To understand how the comprehensive molecular profile translates into drug sensitivity or resistance, two ex vivo, single-cell resolution drug response profiling technologies are included. Pharmacoscopy (Snijder et al., 2017; Vladimer et al., 2017) focuses on cancer-cell-specific drug effectiveness using cell death as a readout, while 4i (iterative indirect immunofluorescence imaging) Drug Response Profiling (Gut et al., 2018) maps the changes in proliferation or survival signaling pathways upon drug treatment, using a multiplexed readout of cancer-relevant molecular markers. Both assays screen a cancer-type-specific set of approved or promising off-label cancer drugs, alone or in combination. Finally, TuPro includes bulk RNA sequencing and targeted DNA sequencing of the tumor and of blood-derived cell-free DNA cfDNA). These well-established cancer-profiling molecular approaches enable the comparison to existing large-scale cohort studies, to leverage their information for patients included in the TuPro study. The unique combination of TuPro technologies overcomes the limitations of current genetic-centric personalized medicine options by providing complementary biomarker data across multiple biological levels and offering a holistic view of a tumor's biology for each individual patient.
[Irmisch et al., Cancer Cell, 2021]
TuPro includes two emerging clinical diagnostic approaches, i.e., tests that are not yet standard in cancer diagnostics, and seven exploratory profiling technologies in the TuPro study. Single-cell genomics approaches (scRNA [Papalexi and Satija, 2018] and scDNA [Kuipers et al., 2020]) generate a high-resolution map of the tumor microenvironment, characterize tumor cell heterogeneity, establish each tumor's evolutionary history, and take advantage of insights into cancer genomics and transcriptomics acquired over the past decades. In addition, bulk (DIA-MS) proteotyping (Gillet et al., 2012; Xuan et al., 2020) and single-cell CyTOF (Wagner et al., 2019) protein-based analyses are performed not only to expand on and translate transcriptomic observations but also to assess post-translational modifications affecting proteins involved in signaling pathways. The characterization of the tumor microenvironment is enriched with spatially resolved approaches: digital pathology and imaging mass cytometry (IMC) (Giesen et al., 2014) enable the characterization of cell-cell interactions within the tumor microenvironment by providing quantitative, single-cell, and spatially resolved data. This is of particular value for predicting the success of therapies that depend on direct cell-cell interactions, such as immune checkpoint inhibitors. To understand how the comprehensive molecular profile translates into drug sensitivity or resistance, two ex vivo, single-cell resolution drug response profiling technologies are included. Pharmacoscopy (Snijder et al., 2017; Vladimer et al., 2017) focuses on cancer-cell-specific drug effectiveness using cell death as a readout, while 4i (iterative indirect immunofluorescence imaging) Drug Response Profiling (Gut et al., 2018) maps the changes in proliferation or survival signaling pathways upon drug treatment, using a multiplexed readout of cancer-relevant molecular markers. Both assays screen a cancer-type-specific set of approved or promising off-label cancer drugs, alone or in combination. Finally, TuPro includes bulk RNA sequencing and targeted DNA sequencing of the tumor and of blood-derived cell-free DNA cfDNA). These well-established cancer-profiling molecular approaches enable the comparison to existing large-scale cohort studies, to leverage their information for patients included in the TuPro study. The unique combination of TuPro technologies overcomes the limitations of current genetic-centric personalized medicine options by providing complementary biomarker data across multiple biological levels and offering a holistic view of a tumor's biology for each individual patient.
[Irmisch et al., Cancer Cell, 2021]